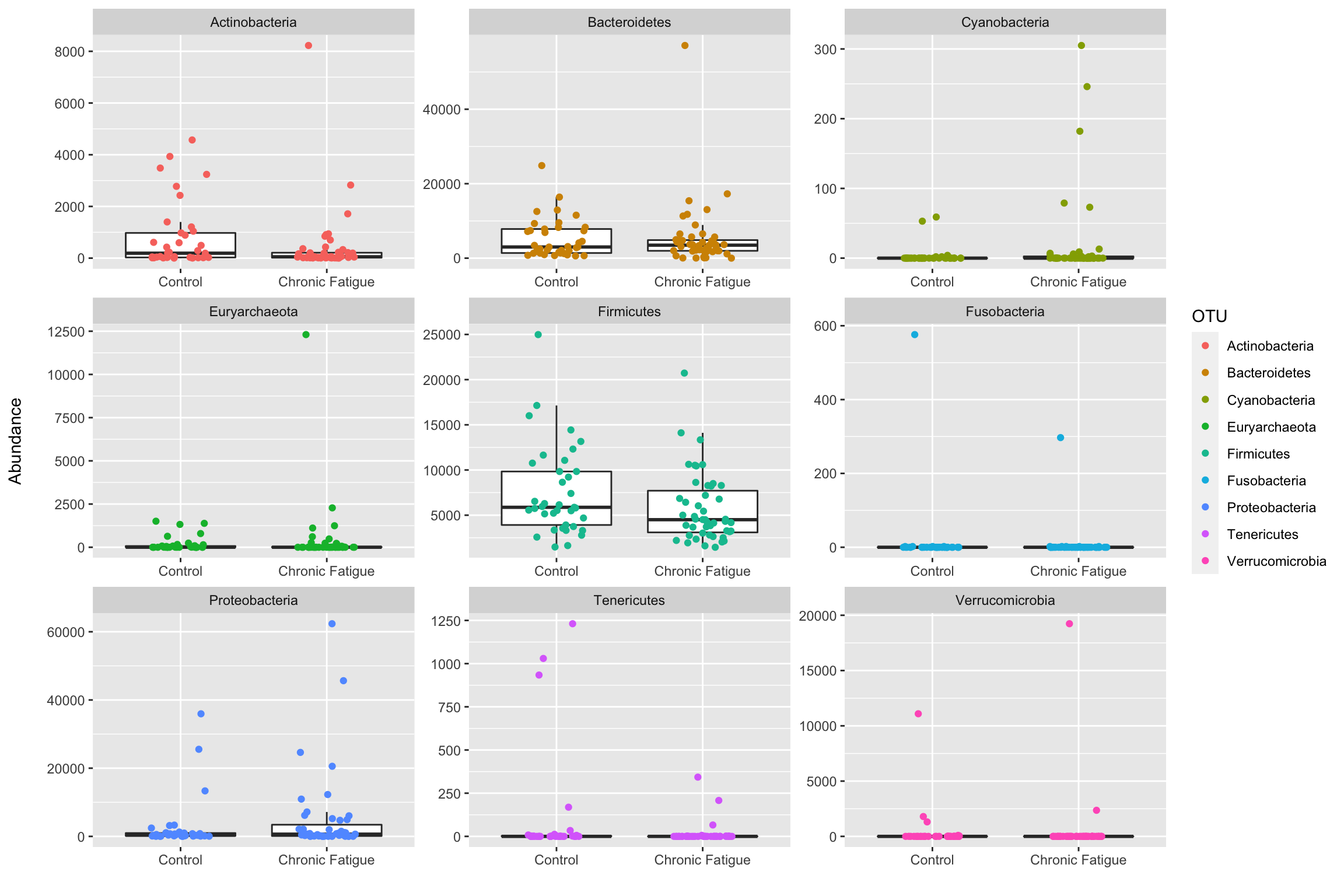
Statitical Method Used to Describe Bacteria Abundance
People am also facing the same challenge on which statistical method to use I am interested in species description and distribution over four agro-ecological zones at different stages of crop. We used this FCM method to compare bacterial abundance measured in four types of crop management systems.

The Distinction Between Absolute Abundances And Relative Abundances As Download Scientific Diagram
Various terms used in this paper are summarized in Table 1.
. Statistical Analysis of Microbiome Data in R by Xia Sun and Chen 2018 is an excellent textbook in this area. 1 Z Ii T Ii Ri Z T Rc T 1 Ri Ri Rc Ii where Ii is the absolute abundance of ISS HAAQ-GFP in each inoculated treatment. The notations described in statistical methods are summarized in.
Bacterial abundance ranged between 018 and 73 x 105 cells ml-1 within the euphotic zone and was generally higher up to 100 meters throughout the study period. Metastats p and q values students t-test. The different diversity characteristics of bacterial and fungal communities.
Cloudy turbid broth indicates greater bacterial population. The term bacterial refers to single-celled organisms bacteria. Useful only if concentration of cells exceeds 1 million per milliliter.
A spectrophotometer doesnt exactly count the bacteria but it does. You can also use some fingerprinting methods to quantify microbial abundance or biomass such as PLFA quantification of lipids even if now Q-PCR is. Then the arithmetic means from different samples are statistically compared.
In petri dishes it is possible to plot the bacterias growth anbsorbances whereas it might be possible to plot incubated times or log of cells versus time. Multiple-correlation analyses using Pearsons method were used to relate archaeal bacterial and fungal abundances. The plate count method or spread plate relies on bacteria growing a colony on a nutrient medium.
Organic integrated with TRC or without tillage RTRC and conventional. A viable cell count allows one to identify the number of actively growingdividing cells in a sample. Robust statistical methods for di erential abundance analysis of metagenomics data Joseph N Paulson Mihai Popy H ector Corrada Bravoz December 17 2011 Abstract This document outlines my 2011-2012 AMSC project for the 663664 course series and in.
With the relative abundance of the Escherichia genus and the absolute abundance of ISS HAAQ-GFP the absolute abundance of the total soil indigenous bacteria was calculated using the following equation. This kind of trend however is known as a sigmoid curve in science. A positive correlation of Firmicutes and 16S rDNA gene abundances across all the 4 protocols was observed Figures 3BE suggesting that indeed bacteria of Firmicutes family represent a good portion of the vagina flora.
Accuracy is affected if cells are not. After calculations are used to detirmine the sizeUsually the procedure is done indirectly with a series of dilutions making it possible to estimate the number of bacteria in the original sample. Absolute abundances tend to be greatly right-skewed in distribution so log-transformation will be useful if statistical methods that assume normal distribution are used.
For those looking for an end-to-end workflow for amplicon data in R I highly recommend Ben Callahans F1000 Research paper Bioconductor Workflow for. Method 2 gave the highest yield of bacterial abundance followed by methods 1 3 and 4 respectively. Up to 10 cash back Pyrosequencing was used to 1 detect and identify bacterial community abundance patterns that described discrete time points of the decomposition process and 2 identify bacterial taxa important for estimating physiological time a timetemperature metric that is often commensurate with minimum post-mortem interval.
The number of bacterial cells in randomly selected fields of view for a given sample 121418. Then we can do the scaling up trick to get the total population size. Estimate number of bacteria by analyzing turbidity with spectrophotometer measures the amount of light passing through a culture.
Transmission of light is inversely proportional to population size of bacteria. Measure how much they interfere with a light beam and based on that we can use a standard curve to decide how many bacteria there are. The 99 of bacteria in the environment that are recalcitrant to culturing have spurred the development of metagenomics a culture-independent approach to sample and characterize microbial genomes.
Up to 24 cash back Most methods of counting are based on indirect or direct counts of tiny samples. This is the most frequently used method. One way we can do this is to use a spectrophotometer.
For the statistical analysis of bacterial abundances relative or absolute the distribution of the data should be considered. The relative abundance of each COG is presented together with significance values likelihood that difference in abundance is due to chance alonecomputed using several statistical procedure. Similarly two other frequently used.
Relative species abundance is a component of biodiversity and is a measure of how common or rare a species is relative to other species in a defined location or community. 454 Pyrosequencing to Describe Microbial Eukaryotic Community Composition Diversity and Relative Abundance. Ri and Rc are the relative.
Instead they should have referred to it as observed abundance. And the bacterial top 15 and fungal top 10 most abundant classes with environmental altitude mean annual air temperature MAT mean annual soil temperature. In offshore waters bacterial production 0401 to 0050 µg C m-3 d-1 abundance 45 to 16 x 105 cells ml-1 and depth of the productive layer decreased from 150 to 75 meters.
A Test for Marine Haptophytes Elianne Egge1 Lucie Bittner24 Tom Andersen3 Stéphane Audic2 Colomban de Vargas2 Bente Edvardsen1 1 University of Oslo Department of Biosciences Marine Biology Oslo Norway 2 CNRS UMR7144 UPMC Station. Relative abundance is the percent composition of an organism of a particular kind relative to the total number of organisms in the area. Bacterial waste products debris in the GIT can be quantified by analysis of microbial structural patterns specific to bacteria.
The arithmetic mean value of bacterial counts obtained under the most favorable conditions was considered as the true estimate of bacterial abundance 18. How Is Bacterial Growth Curve Used. The FCM count ranged from 391 10 8 to 569 10 8 cells g 1 soil for crop field and was 669 10 8 cells g 1 soil for grassland.
Peptidoglycan makes up approximately 90 of gram-positive bacteria cell walls and is also a minor component in gram-negative. There are a variety of ways to enumerate the number of bacteria in a sample. Robust statistical methods for di erential abundance analysis of metagenomics data Joseph N Paulson Mihai Popy H ector Corrada Bravoz May 15 2012 Abstract This document outlines my 2011-2012 AMSC project for the 663664 course series.
One abundant microbial polymer unique to bacteria is the cell wall component peptidoglycan. There are many great resources for conducting microbiome data analysis in R. Massive datasets of metagenomic sequences have been accumulated but analysis of these sequences has focused primarily on the descriptive comparison of the.

Bacterial Abundance Types Bacterial Abundance Types Include Symmetric Download Scientific Diagram
Quantitative Pcr Provides A Simple And Accessible Method For Quantitative Microbiota Profiling
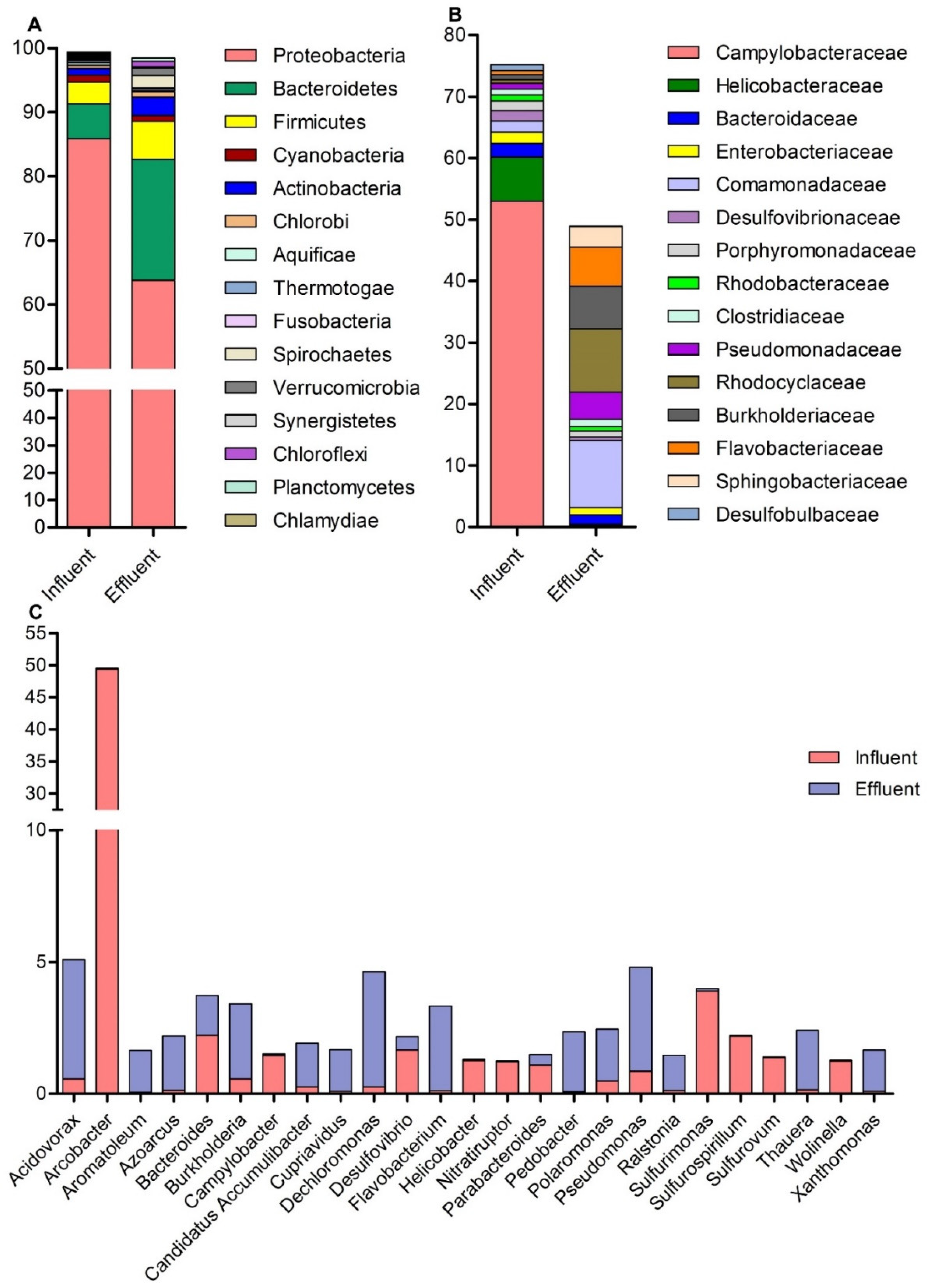
Diversity Free Full Text Analysis Of Microbial Communities And Pathogen Detection In Domestic Sewage Using Metagenomic Sequencing Html
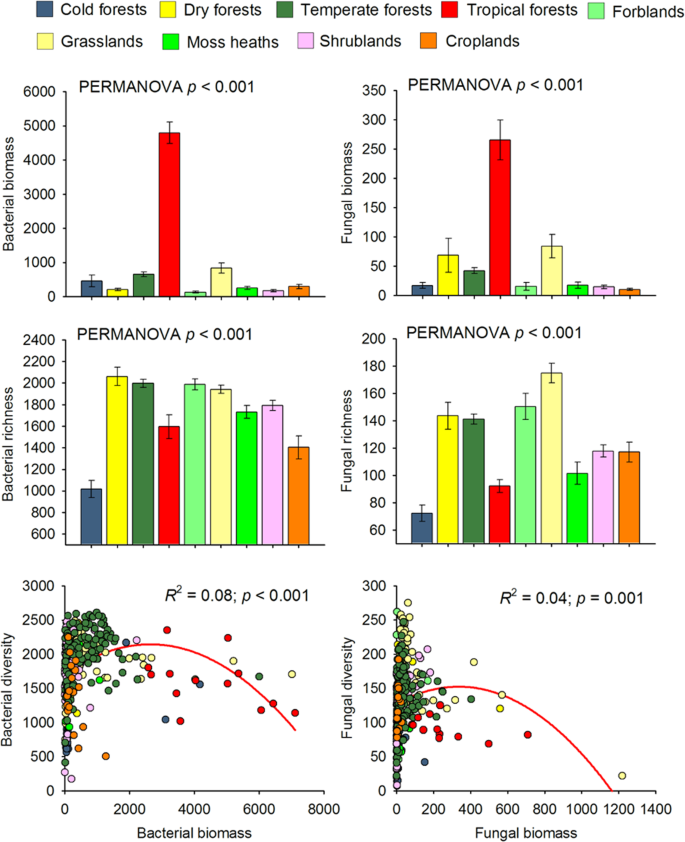
Soil Microbial Diversity Biomass Relationships Are Driven By Soil Carbon Content Across Global Biomes The Isme Journal
A Statistical Model For Describing And Simulating Microbial Community Profiles
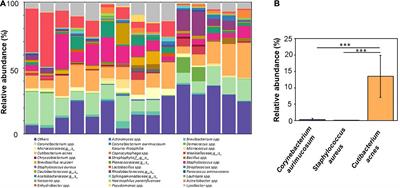
Circulating Antibodies To Skin Bacteria Detected By Serological Lateral Flow Immunoassays Differentially Correlated With Bacterial Abundance Microbiology Frontiers
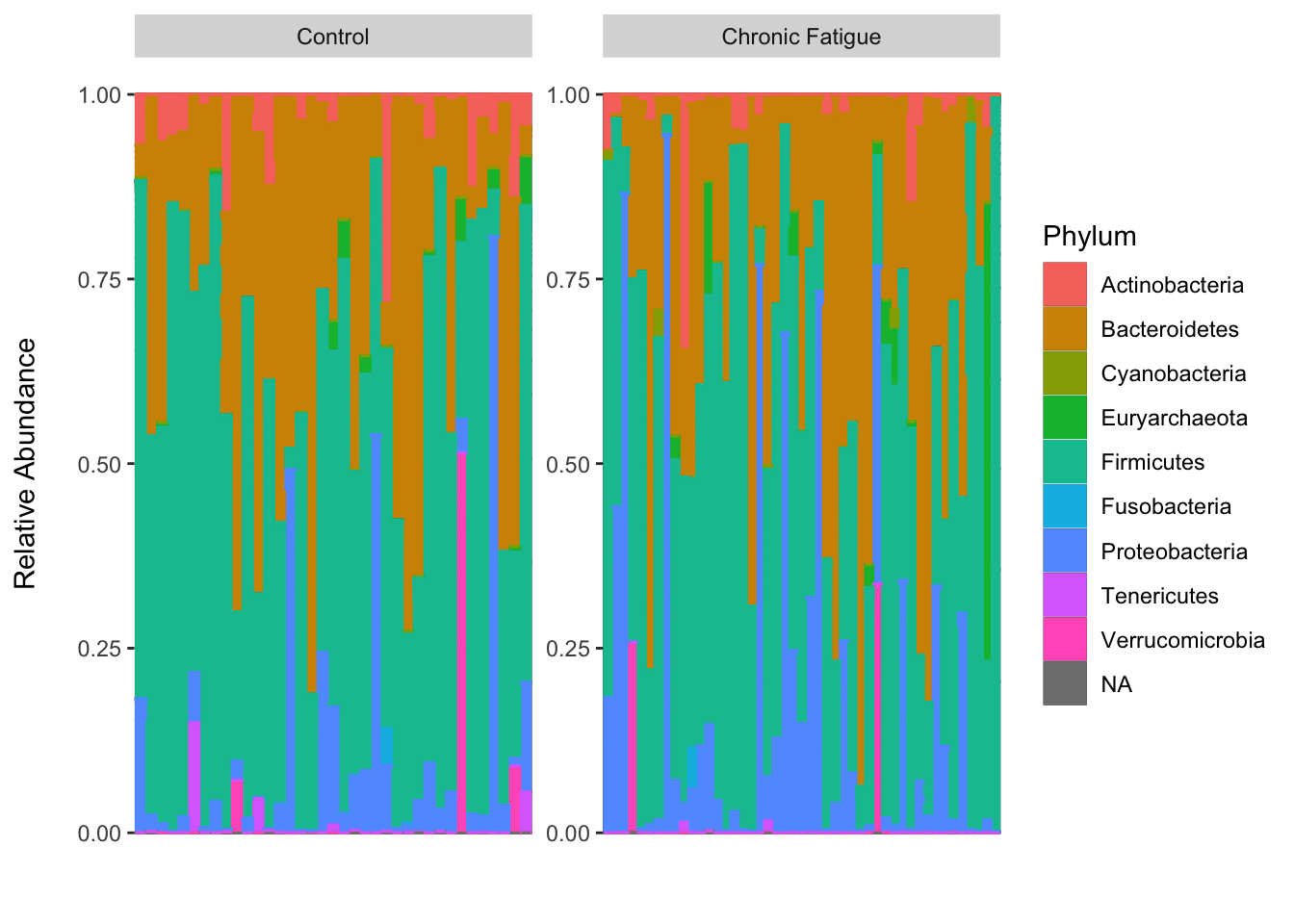
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
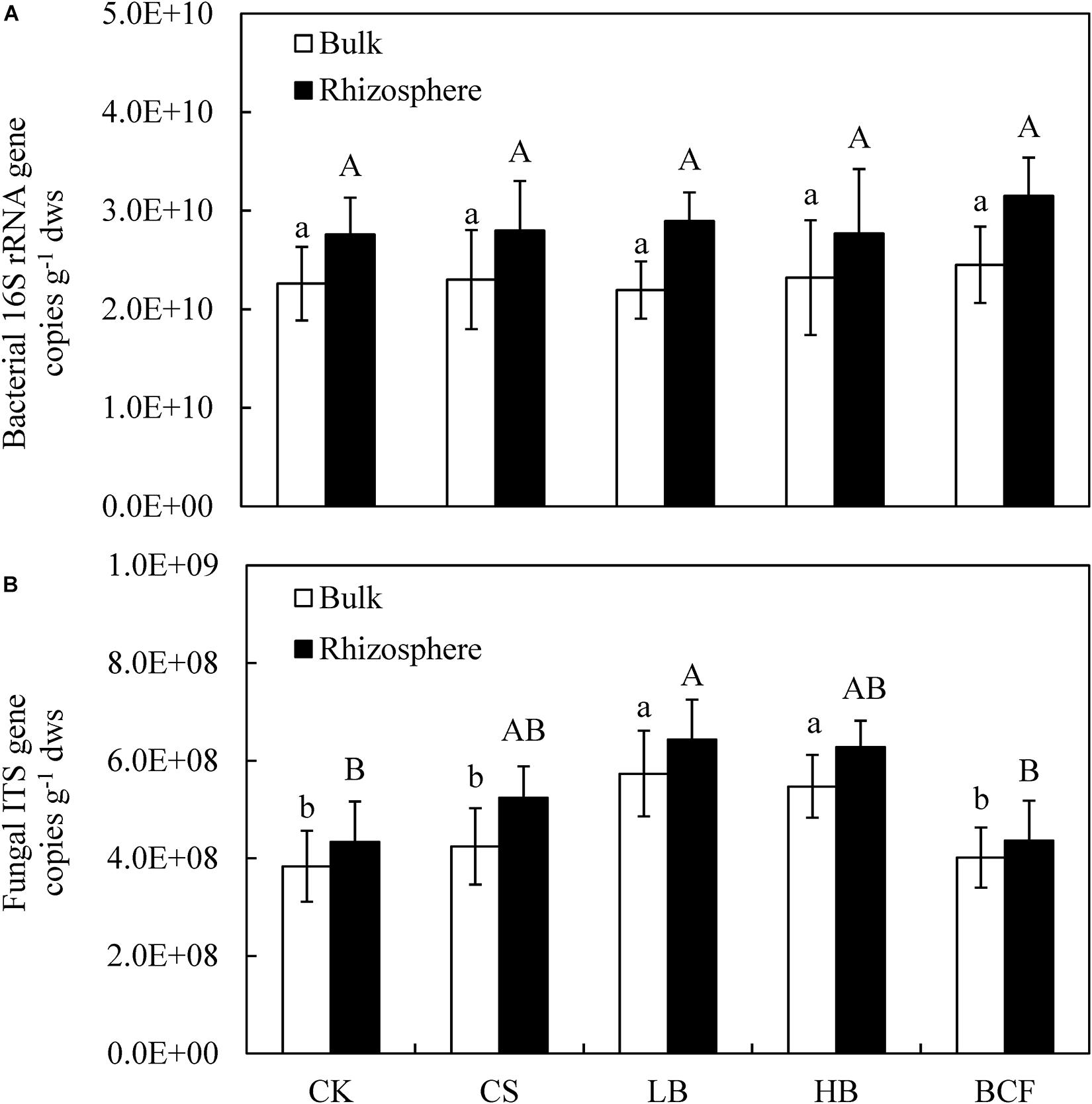
Frontiers Different Responses Of Soil Bacterial And Fungal Communities To 3 Years Of Biochar Amendment In An Alkaline Soybean Soil Microbiology
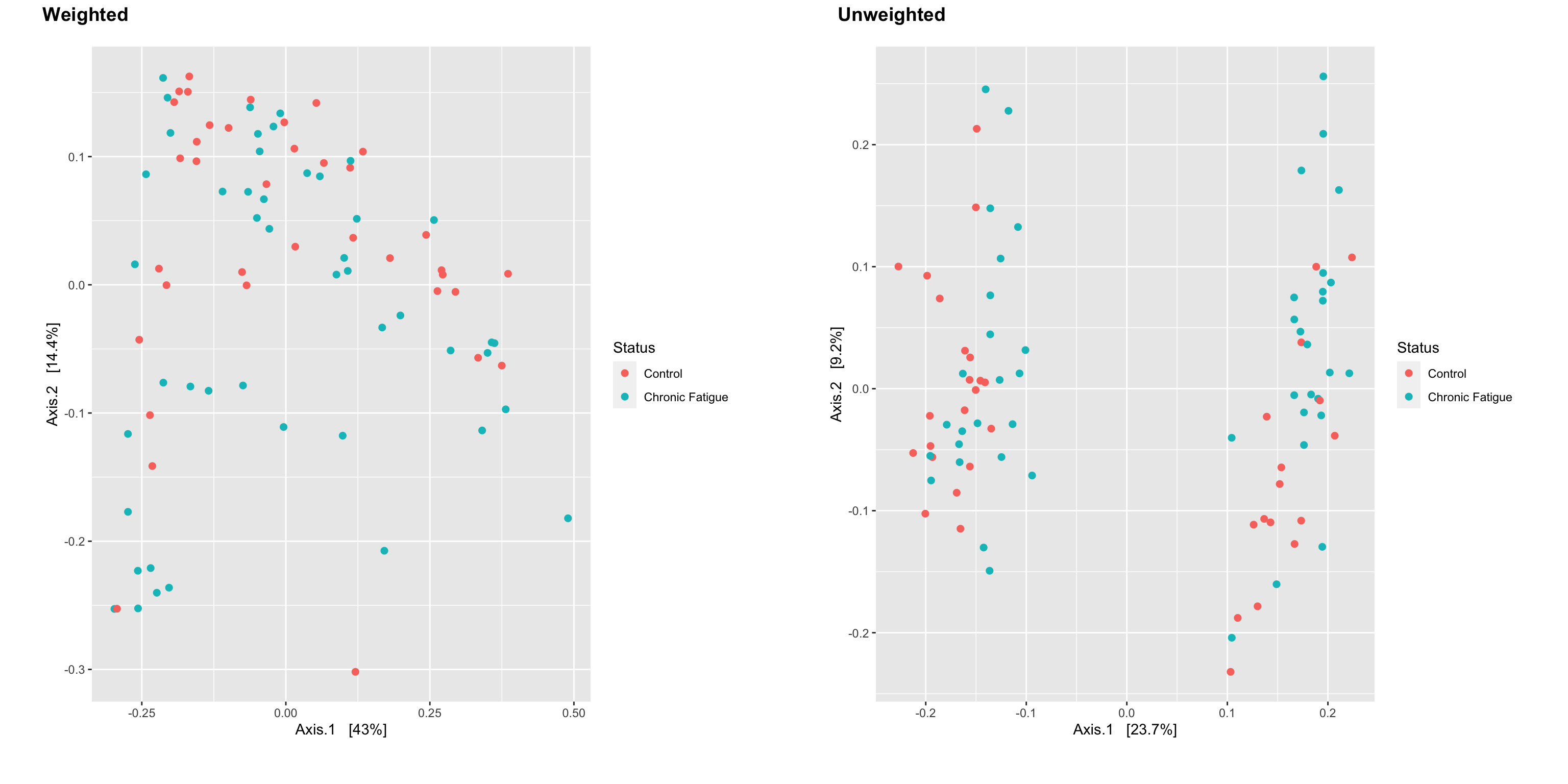
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
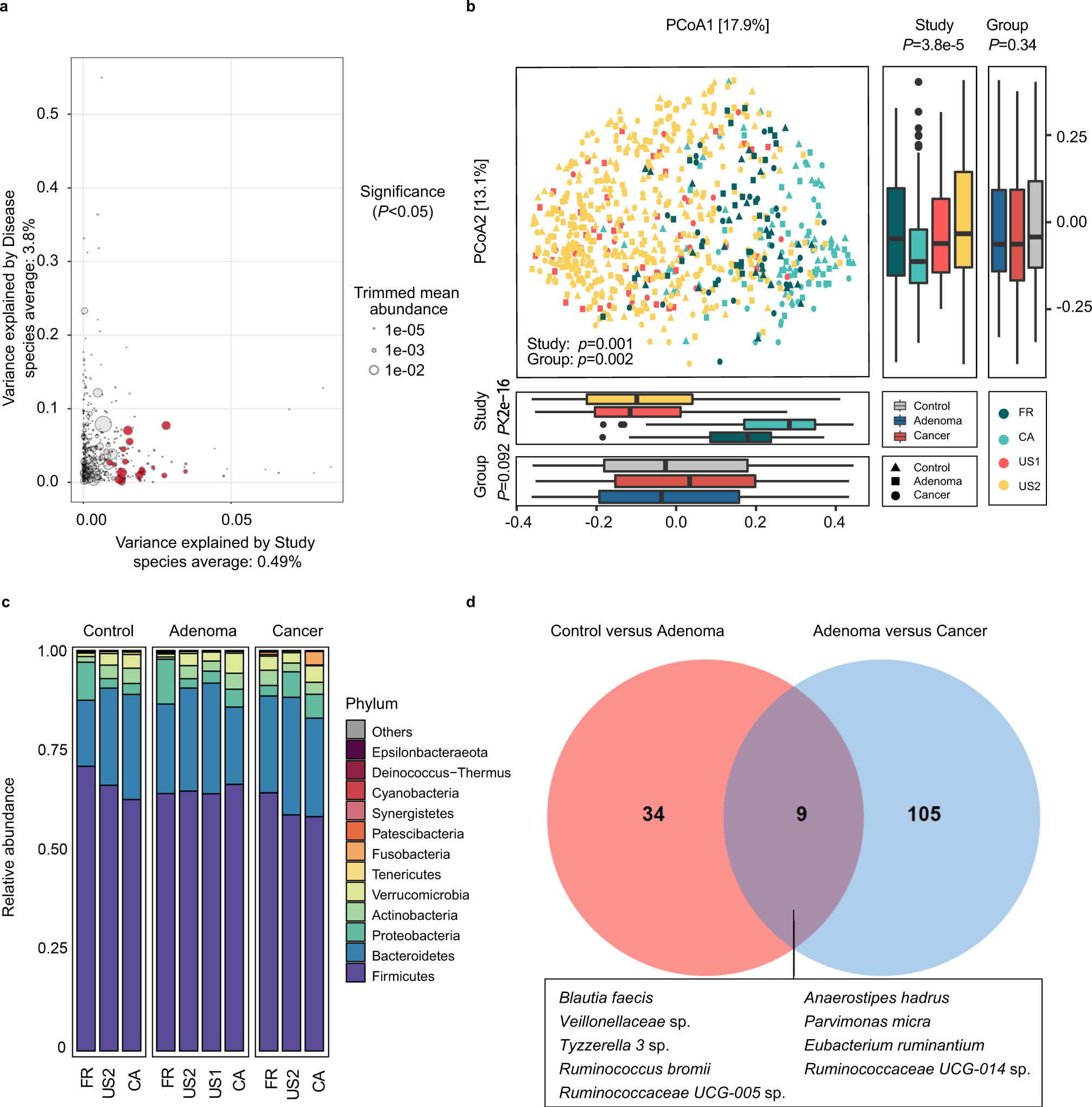
Identification Of Microbial Markers Across Populations In Early Detection Of Colorectal Cancer Nature Communications

High Viral Abundance And Low Diversity Are Associated With Increased Crispr Cas Prevalence Across Microbial Ecosystems Sciencedirect
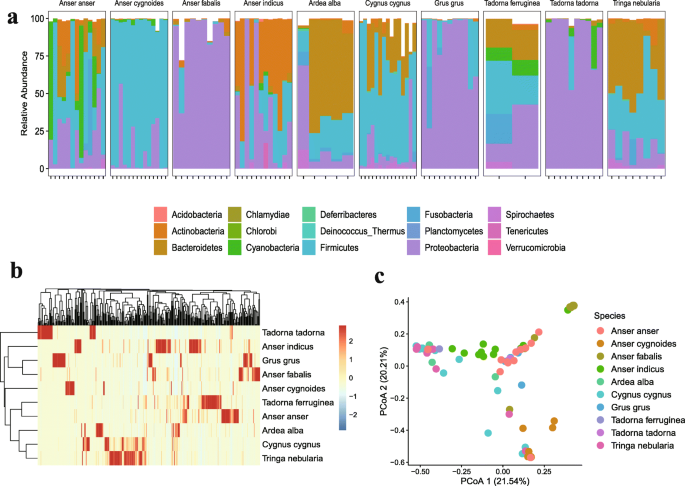
Metagenomic Analysis Reveals The Microbiome And Resistome In Migratory Birds Springerlink
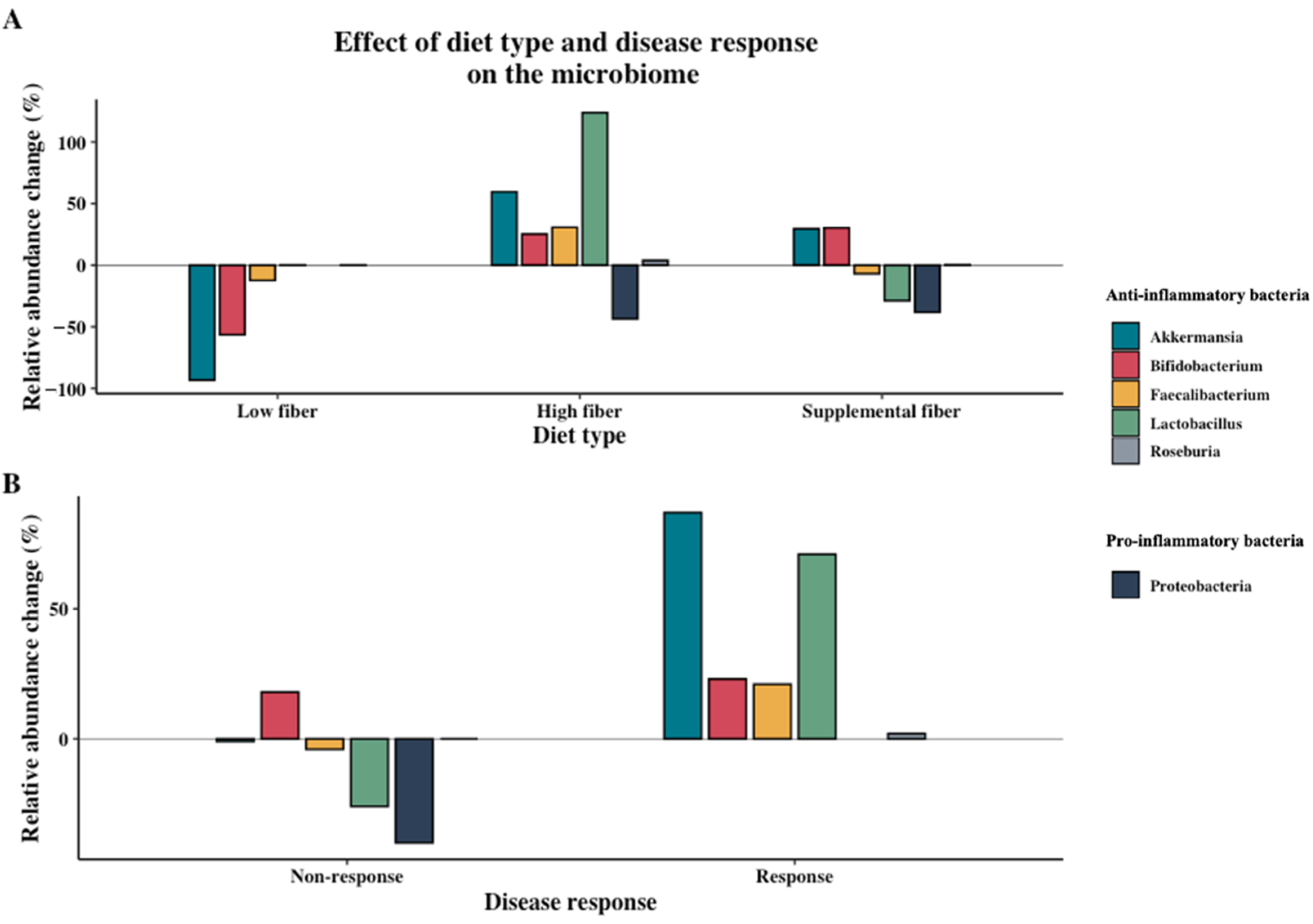
Nutrients Free Full Text The Effect Of Dietary Interventions On Chronic Inflammatory Diseases In Relation To The Microbiome A Systematic Review Html

The Distinction Between Absolute Abundances And Relative Abundances As Download Scientific Diagram
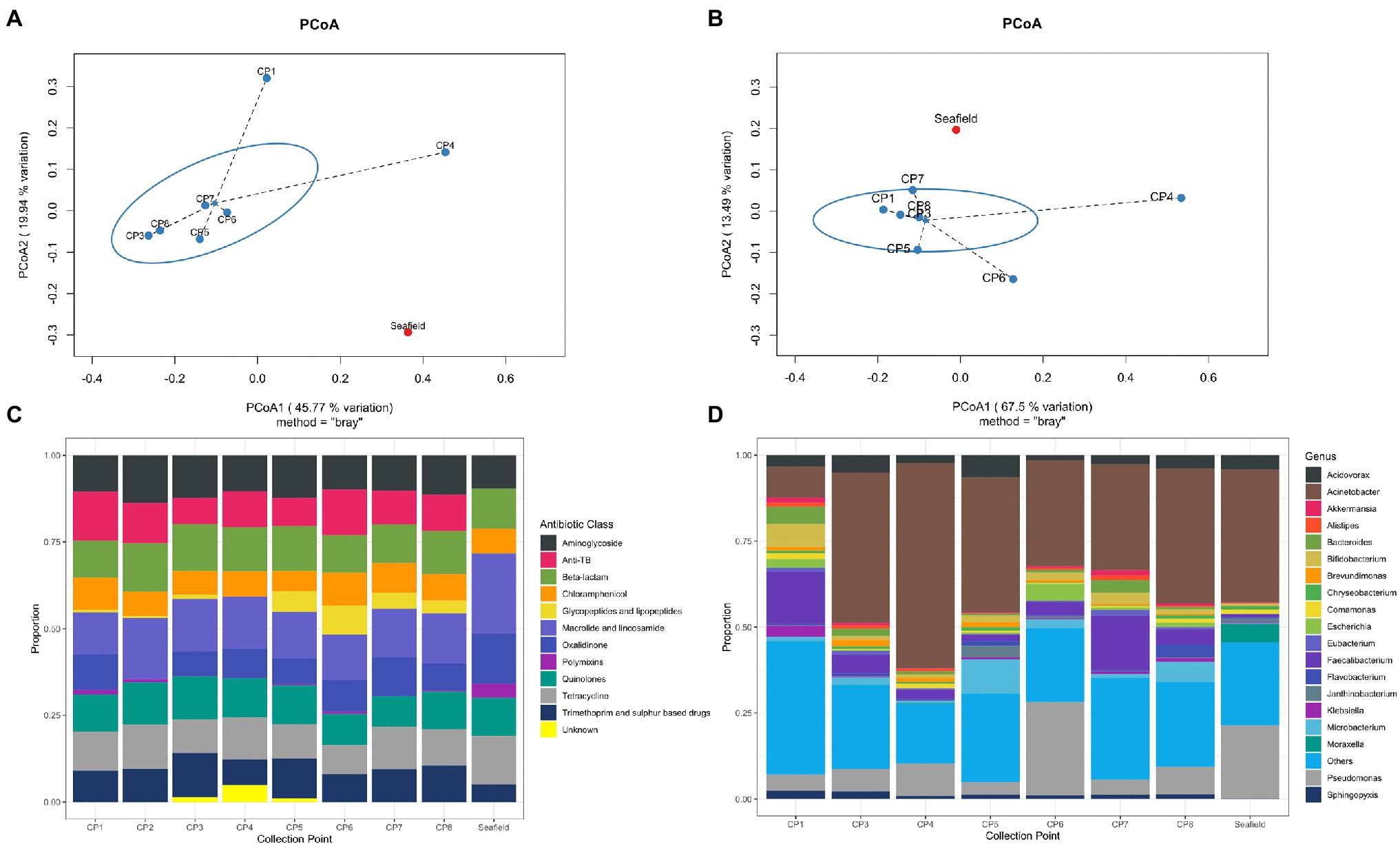
Frontiers Secrets Of The Hospital Underbelly Patterns Of Abundance Of Antimicrobial Resistance Genes In Hospital Wastewater Vary By Specific Antimicrobial And Bacterial Family Microbiology

Introduction To The Statistical Analysis Of Microbiome Data In R Academic
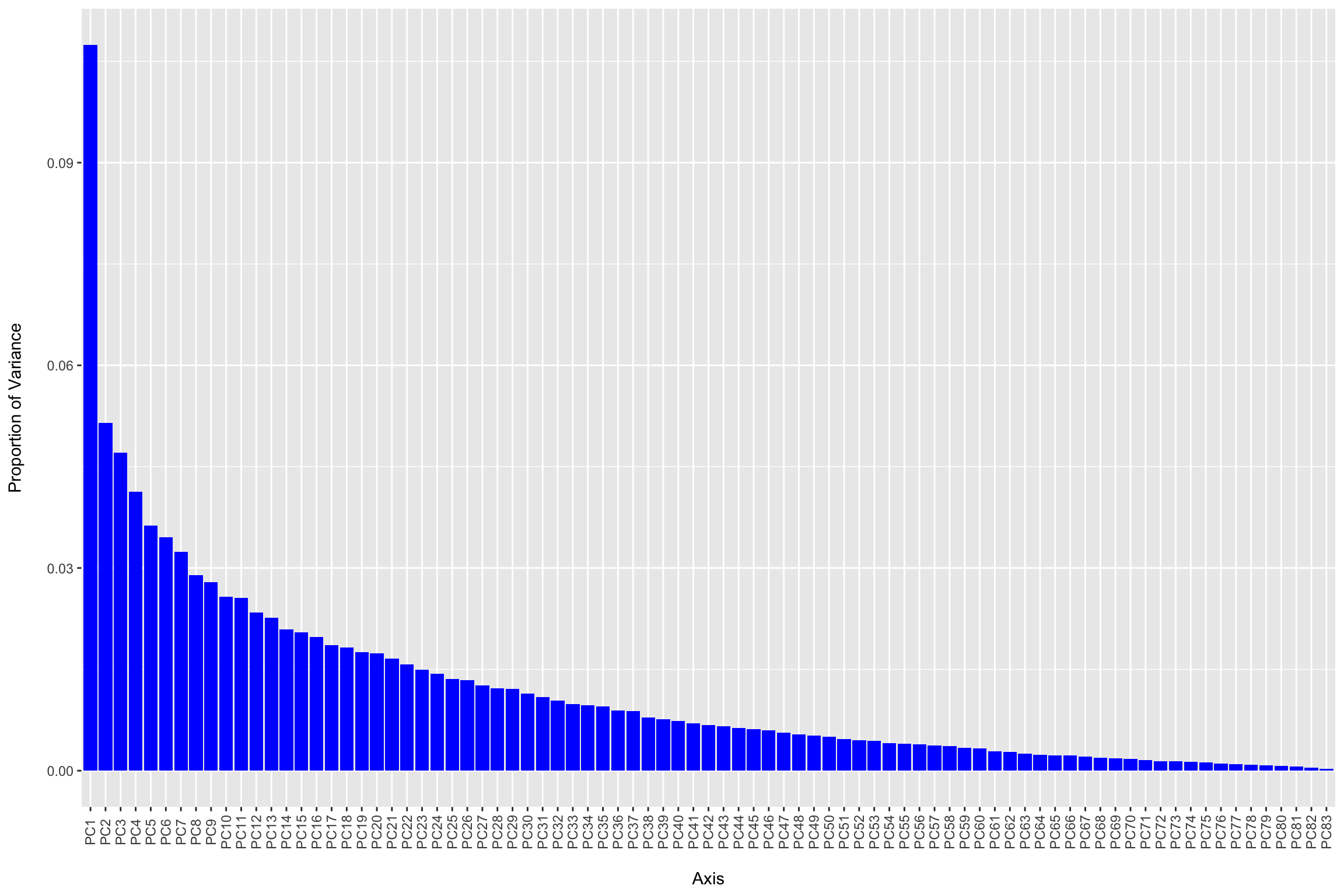
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
Multi Kingdom Microbiota Analyses Identify Bacterial Fungal Interactions And Biomarkers Of Colorectal Cancer Across Cohorts Nature Microbiology
Comments
Post a Comment